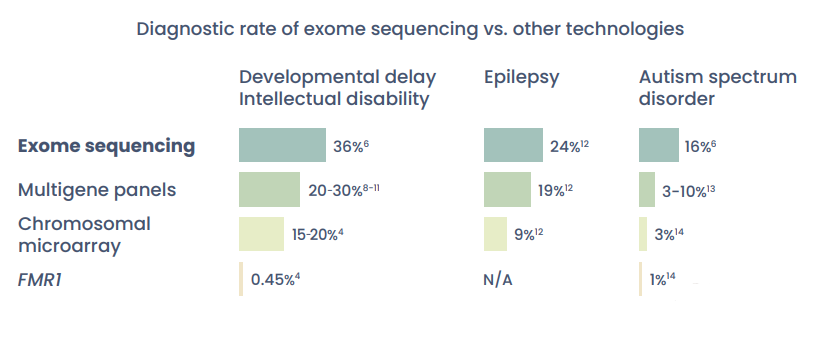
Whole exome sequencing is more likely to deliver a genetic diagnosis than multigene panels or chromosomal microarray.

An earlier genetic diagnosis is proven to:4,5

American College of Medical Genetics and Genomics recommends exome or genome as a first-tier test for developmental delay, intellectual disability, and congenital anomalies.15
National Society of Genetic Counselors recommends exome or genome sequencing for all individuals with unexplained epilepsy. This guideline is endorsed by the American Epilepsy Society.16
Choosing exome sequencing can shorten the journey to a diagnosis, resulting in updated medical management and reduced healthcare expenses.
The diagnostic rate of exome testing is 2x greater than chromosomal microarray.4,6
23% of patients diagnosed via exome testing would not have received a diagnosis with a genetic panel.7
XomeDx® Plus consists of concurrent evaluation of the exome and mitochondrial genome using two separate assays. Separate result reports will be issued for the exome analysis and the mitochondrial genome analysis. XomeDx® Plus is best suited for individuals with clinical features suggesting a mitochondrial disorder. Select trio, duo, or proband.
Turnaround time: 6 weeks*
*Turnaround times are estimates and begin once the sample(s) begin processing at the GeneDx lab and could be extended in situations outside GeneDx’s control.
†Fictionalized case study for illustrative purposes only
References