Accelerating diagnosis, improving NICU outcomes
Lorem ipsum dolor sit amet, consectetur adipiscing elit. Suspendisse varius enim in eros elementum tristique. Duis cursus, mi quis viverra ornare, eros dolor interdum nulla, ut commodo diam libero vitae erat. Aenean faucibus nibh et justo cursus id rutrum lorem imperdiet. Nunc ut sem vitae risus tristique posuere.

Of infants admitted to level IV NICUs admissions are likely eligible for rapid genome sequencing1.
Of eligible patients receive rapid testing2. For these critically ill infants, every moment matters. Rapid genome sequencing can provide an early and precise diagnosis–enabling timely interventions, reducing hospital stays, and improving outcomes3,4.
Current NICU protocols overlook patients
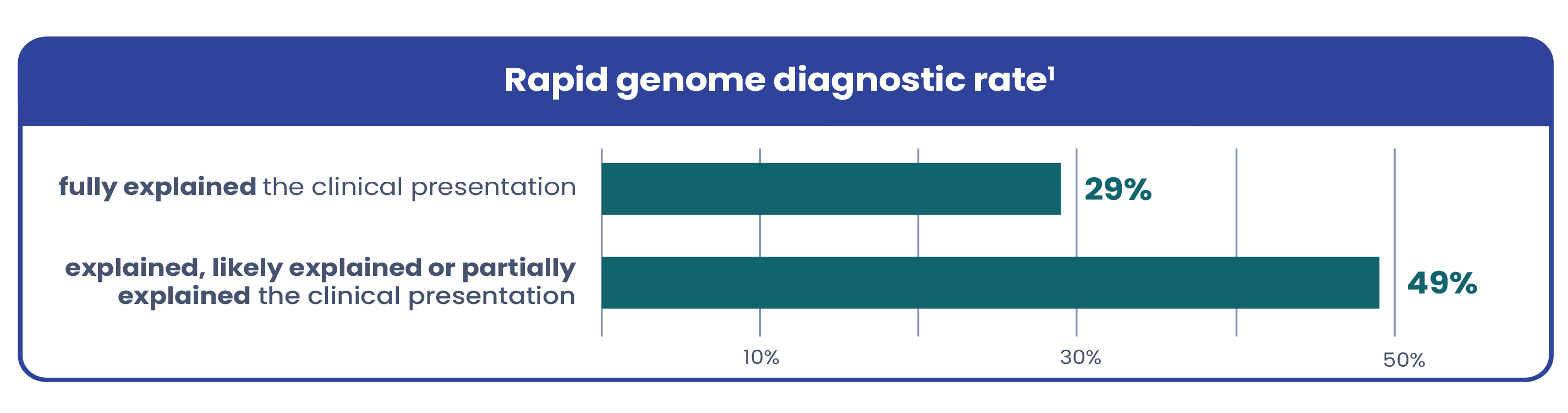
Due to variable and complex presentations, genetic conditions in critically ill infants are often hard to recognize based on symptoms alone. Current testing workflows are often too narrow, leaving many patients undiagnosed. A recent study in The American Journal of Human Genetics found that among the infants who received a genetic diagnosis via rapid genome with a broad testing protocol1:
Infants could be up to 9x more likely to receive a genetic diagnosis when using a broad testing protocol compared to a conventional workflow1
Testing all infants whose clinical condition is not fully explained by trauma, infection, or prematurity leads to more diagnoses and maintains a high diagnostic yield1.
Genome sequencing is recommended as a first-line test
- The International Precision Child Health Partnership (IPCHiP) recommends genome or exome as a first-line test for NICU patients with unexplained hypotonia6.
- The American College of Medical Genetics and Genomics (ACMG) recommends genome or exome as a first-line test for developmental delay, intellectual disability, and congenital anomalies7.
- The National Society of Genetic Counselors (NSGC) recommends genome or exome as a first-line test for all individuals with unexplained epilepsy and this guideline is endorsed by the American Epilepsy Society (AES)8.
- The American Academy of Pediatrics (AAP) recommends ordering exome and genome as a first-line tests for children with global developmental delays and intellectual disabilities9.


Real stories. Real diagnoses.
Infinity is shaped by the data of real people and real cases – and it’s changing what’s possible in rare disease care. It helps us see more clearly, diagnose more confidently, and discover insights that lead to better answers for more families.
Helpful resources
No-charge reanalysis service
GeneDx provides one reanalysis at no additional charge per genome order, recommended at least one year after the original analysis to maximize gene discovery and to identify new patient phenotypes that may inform reporting (especially in the NICU population).
GeneDx Infinity™
Infinity is powered by data from over 2.5 million tests – informed by nearly 1 million exomes and genomes and more than 7 million phenotypic datapoints – to help you uncover answers faster and fuel the discovery of life-changing treatments.
Rapid genome ordering guide
Access the Rapid Genome Ordering Guide for NICU teams—featuring step-by-step ordering instructions, specimen guidance, and contact pathways for the dedicated Xpress support team.